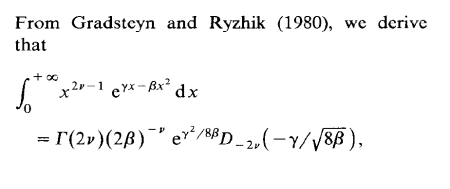 One of my students in my MCMC course at ENSAE seems to specialise into spotting typos in the Monte Carlo Statistical Methods book as he found an issue in every problem he solved! He even went back to a 1991 paper of mine on Inverse Normal distributions, inspired from a discussion with an astronomer, Caroline Soubiran, and my two colleagues, Gilles Celeux and Jean Diebolt. The above derivation from the massive Gradsteyn and Ryzhik (which I discovered thanks to Mary Ellen Bock when arriving in Purdue) is indeed incorrect as the final term should be the square root of 2β rather than 8β. However, this typo does not impact the normalising constant of the density, K(α,μ,τ), unless I am further confused.
One of my students in my MCMC course at ENSAE seems to specialise into spotting typos in the Monte Carlo Statistical Methods book as he found an issue in every problem he solved! He even went back to a 1991 paper of mine on Inverse Normal distributions, inspired from a discussion with an astronomer, Caroline Soubiran, and my two colleagues, Gilles Celeux and Jean Diebolt. The above derivation from the massive Gradsteyn and Ryzhik (which I discovered thanks to Mary Ellen Bock when arriving in Purdue) is indeed incorrect as the final term should be the square root of 2β rather than 8β. However, this typo does not impact the normalising constant of the density, K(α,μ,τ), unless I am further confused.
Archive for confluent hypergeometric function
I thought I did make a mistake but I was wrong…
Posted in Books, Kids, Statistics with tags Charles M. Schulz, confluent hypergeometric function, course, ENSAE, exercises, Gradsteyn, inverse normal distribution, MCMC, mixtures, Monte Carlo Statistical Methods, Peanuts, Ryzhik, typos on November 14, 2018 by xi'anLe Monde puzzle [#743]
Posted in R, Statistics with tags confluent hypergeometric function, R on October 12, 2011 by xi'an As Le Monde weekend has yet again changed its format (with so much more advertisements for luxurious items that I sometimes wonder whether or not this is the weekend edition of Le Monde!], it took me a while to locate the mathematical puzzle. The good news is there now is a science&techno leaflet with, at the end, the math puzzle! (Sorry, this is of no relevance for anyone but me!)
As Le Monde weekend has yet again changed its format (with so much more advertisements for luxurious items that I sometimes wonder whether or not this is the weekend edition of Le Monde!], it took me a while to locate the mathematical puzzle. The good news is there now is a science&techno leaflet with, at the end, the math puzzle! (Sorry, this is of no relevance for anyone but me!)
The problem of last week is loosely connected with random walks on a grid. Given a nxm grid, consider all the shortest length paths linking (0,0) with (n,m). They all are of length n+m and can be represented as a sequence of l‘s and d‘s (for left and down). The question amounts to compute the probability that two of those paths intersect “in the middle”, i.e. after (n+m)/2 steps if (n+m) is even, or share the edge from the (n+m-1)/2 node to the (n+m+1)/2 node.
When (n+m) is even, if the meeting node is (i,j), with i+j=(n+m)/2, there are
moves from (0,0) to (i,j) and then again
moves from (i,j) to (n,m) (since the total number of down moves is n). The number of paths going from (0,0) to (n,m) through (i,j) is therefore
and the probability of meeting in (i,j) is
hence the probability of meeting somewhere is
whose numerator is equal to
according to Wolfram alpha when n>m…. And to
otherwise. Nothing we can compute in R, I am afraid. (Feel free to process the case when n+m is odd as an exercise!)
Now, the original problem involved a random choice of direction (left and down) at each node (if any), meaning all paths do not have the same probability. So here is a (plain) R code aiming at approximating the probability:
N=10^3
n=7
m=11
genepath=function(n,m){
path=matrix(0,ncol=2,nrow=n+m+1)
for (t in 2:(n+m+1)){
if (max(path[,1]-n,path[,2]-m)<0){
path[t,]=path[t-1,]+sample(c(0,1),2)
}else{
path[t,]=path[t-1,]+c((path[t-1,1]<n),(path[t-1,2]<m))
}}
path
}
cross=0
dega=hcl(h = seq(360, 340, length = N)+100,
l = seq(90, 10, length = N))
par(mar=c(0,0,0,0))
plot(seq(0,n,le=100),seq(0,m,le=100),axes=FALSE,col="white",
xlab="",ylab="")
for (a in 1:N){
path1=genepath(n,m)
path2=genepath(n,m)
lww=.3 #width of path
#signals intersections
if ((path1[1+(n+m)/2,1]==path2[1+(n+m)/2,1])&&
(path1[1+(n+m)/2,2]==path2[1+(n+m)/2,2])){
points(jitter(path1[1+(n+m)/2,1]),jitter(path1[1+(n+m)/2,2]),
col="sienna2",pch=19)
cross=cross+1
lww=1}
#separates paths
drift=jitter(path1-path2)
lines(path2+drift,col=dega[a],lwd=lww)
lines(path1-drift,col=dega[a],lwd=lww)
}
cross/N
